CUHK
News Centre
CUHK Researchers Unveil a Novel Competition Strategy that May Fuel Plant Cells for a Sustainable Development of Crops
Rapid climate change is contributing to decreased productivity in crops. A balance in nutrition and energy recycling is critical for plants to survive under different stress conditions. A research team from the School of Life Sciences at The Chinese University of Hong Kong (CUHK) has unveiled a novel competitive mechanism of autophagy in the model plant Arabidopsis thaliana. This work provides an important basis for improving the nutrient and energy efficiency in crops to achieve a sustainable development. The results have been recently published in Autophagy, a top journal in this field.
Autophagy helps to recycle garbage in cells
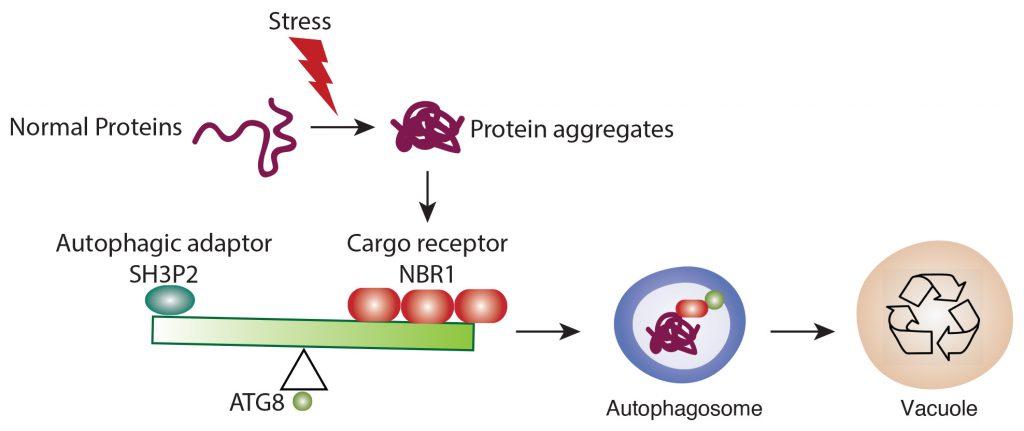
Autophagy, a “self-eating” metabolism pathway conserved in all eukaryote cells, plays a key role in sorting garbage in the cell. For example, when the nutrient supply is limited or the proteins and organelles are damaged, the cells will produce a double-membrane structure called autophagosomes in the cytoplasm. The autophagosome, like a recycling vehicle, will carry the cargo (the garbage) and finally transport it into the vacuole, the recycling centre in the cell. Finally, the cargo will be degraded and released into small building blocks such as amino acids and energy, to fuel cells. Then, it will be recycled to synthesise new proteins. When autophagy does not function properly, the waste cannot be deposited into the recycling centre and accumulates in the cell. Meanwhile, the accumulated proteins in the cell become toxic and finally lead to cell death and disease. Therefore, autophagy is essential in aging and many other developmental processes of diseases.
The discovery of autophagy by a Japanese biologist won the Nobel Prize in Physiology or Medicine in 2016. Yet, there are still many fundamental questions unsolved in autophagy research. Among more than 40 autophagy-related (ATG) proteins, the ATG8 family is a key regulator in the process of biogenesis. Previous research showed that most of the interaction between adaptors ATG8 and receptors NBRI are fueled by an amino acid sequence called ATG8 family interacting motif (AIM). However, the mechanism of how ATG8 protein distinguishes different garbage has been unclear.
An atypical interaction found in the plant model Arabidopsis
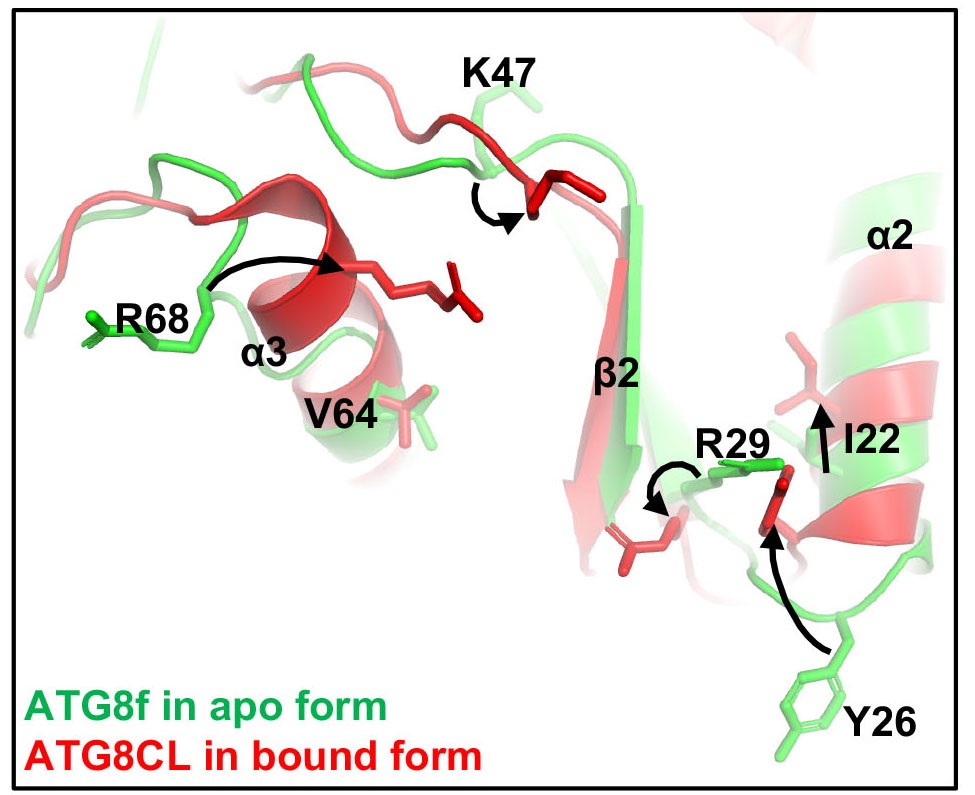
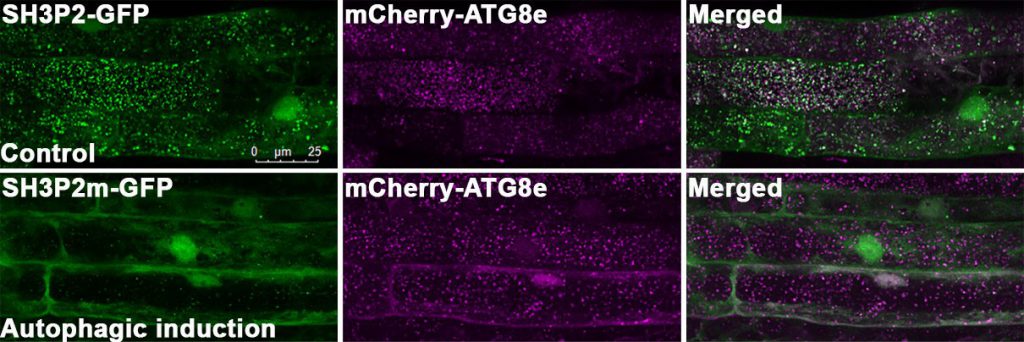
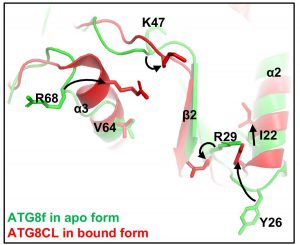
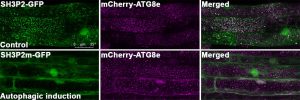
In this study, Professor Xiaohong ZHUANG, Assistant Professor, School of Life Sciences at CUHK, collaborated with Professor Kam Bo WONG, a structural biologist and Director of the School, and Professor Liwen JIANG, Choh-Ming Li Professor of Life Sciences to firstly demonstrated a competition mode in plant selective autophagy at the structural level. The research team studied the ATG8 protein structure in the model plant system Arabidopsis thaliana, and uncovered an atypical ATG8 protein interaction pattern. Among the three SH3-domain-containing proteins in Arabidopsis, analysis revealed that ATG8 only interacts specifically with SH3P2 and utilises a unique interface to recognise it. In addition, the amounts of receptor proteins NRBI can affect the interaction affinity between ATG8 and SH3P2. It is revealed that this competitive mechanism may enable the cells to efficiently make a decision for cargo selection to meet the demands of their nutritional and energy needs. That means more or less garbage will be delivered to the vacuole subject to the needs.
Professor Zhuang remarks, “The new knowledge acquired through this research on autophagy in plants will help in searching novel stress responsive targets for regulating autophagy, thus allowing plants to cope better with stress like nutrient deficiency and drought conditions. In future, our research focus will continue to explore the autophagy network in plants for improving the productivity of crops for a sustainable agriculture.”
Other team members include Dr. Shuangli SUN, Dr. Kin Pan CHUNG, Dr. Ka-Ming LEE, and postgraduate students Lanlan FENG, Hayley Hei-Yin CHEUNG, Mengqian LUO, Kaike REN and Kai Ching LAW.
About Professor Xiaohong ZHUANG
Professor Xiaohong ZHUANG, a plant cell biologist in the School of Life Sciences, has made several key contributions to plant autophagy research. Her work has been published in more than 30 publications, including Proceedings of the National Academy of Sciences of the USA, Autophagy, The Plant Cell, Plant Physiology, and Trends in Plant Science. She was the recipient of the Early Career Scheme 2020-2021 by the Research Grants Council of Hong Kong (RGC), and received funding from the National Natural Science Foundation of China/RGC Joint Research Scheme 2020/2021.
The full text of journal paper can be found at https://doi.org/10.1080/15548627.2021.1976965.
Mechanistic insights into an atypical interaction between ATG8 and SH3P2 in Arabidopsis thaliana. Autophagy. 2021.

The research team is led by Professor Xiaohong ZHUANG (third from the right), in collaboration with Professor Kam Bo WONG(third from the left) and Professor Liwen JIANG (first from the right).

Professor Xiaohong ZHUANG and her PhD student Lanlan FENG in preparing the young Arabidopsis plants in the plant growth room.
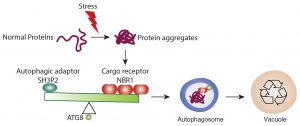
The amounts of receptor proteins NRBI can affect the interaction affinity between ATG8 and SH3P2. This competitive mechanism may enable the cells to efficiently make a decision for cargo selection.